Research
Causal Mediation Analysis with Longitudinal Mediators & Competing Risks
My practicum project in the Valeri Lab focuses on extending the CMAverse R package. I am developing a user‑friendly command that automates causal mediation analysis when (i) mediators are measured repeatedly over time and (ii) both mediators and outcomes are subject to competing events such as death.
Diseases of aging—e.g. dementia—often face the competing risk of mortality. Ignoring this alters estimates of indirect effects. Our command will provide path‑specific effect estimates that remain causally interpretable under standard identifiability assumptions (exchangeability, positivity, consistency). In this project, I aim to develop, implement, and validate (via simulation) an R function that estimates path‑specific effects with longitudinal mediators and competing risks, publish a manual and Github tutorial for this function to assist collaborators and researchers, and apply the new function to simulated datasets.
Functional Data Analysis of Mitochondrial Morphology
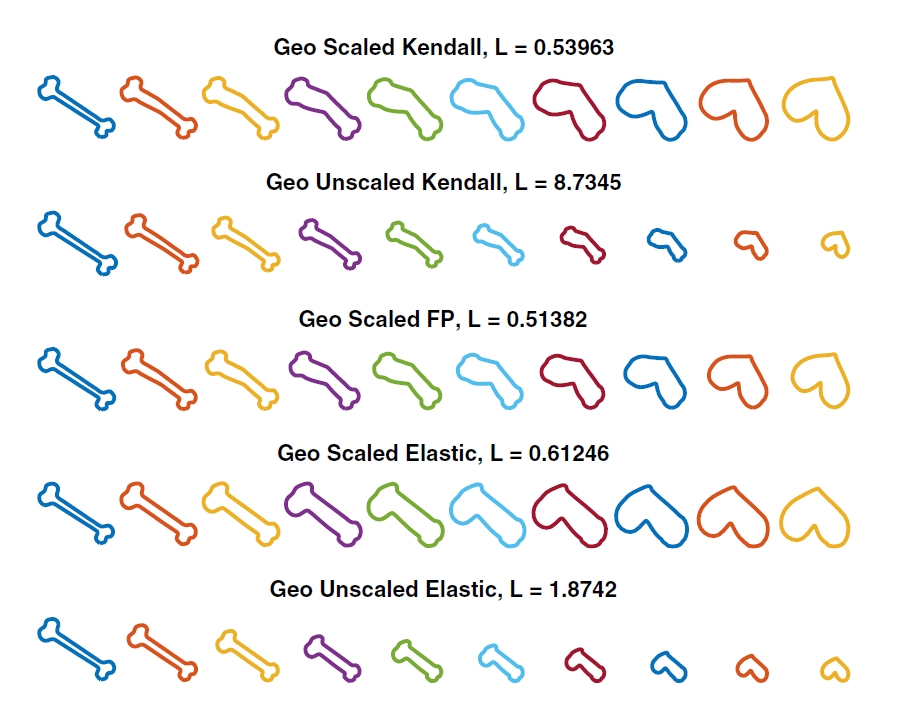
Under Prof. Todd Ogden, I apply functional data analysis tools (MHD, fdasrvf)
to 3‑D mitochondrial shape trajectories in running vs. sedentary mice. We quantify shape
variability along geodesic paths and relate morphological features to metabolic phenotypes.
The figure illustrates geodesic deformation between hypothetical shapes (adapted from
Zhang et al., "Nonparametric k‑sample test on shape spaces with applications to mitochondrial shape analysis", 2022).
Avian Cranial Evolution
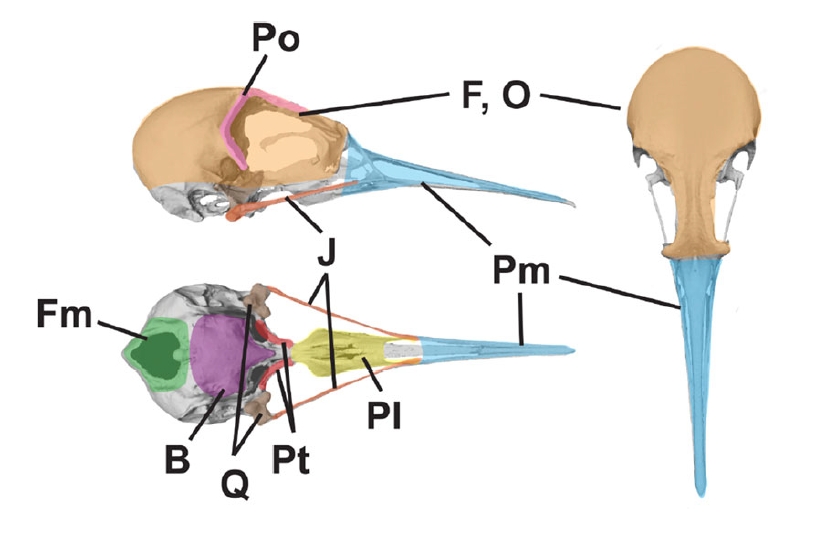
I worked with Rossy Natale, Prof. Graham Slater, and Prof. John Bates to examine the semi‑independent evolution of bird beaks and braincases using high-density 3-D landmarks from shorebirds (order Charadriiformes). We compared 11 alternative “modularity” models. We found that the beak has followed its own semi-independent evolutionary path while remaining moderately correlated with the rest of the skull. This research project has been presented at the 2022 American Ornithological Society Annual Conference in Puerto Rico.